MRC Laboratory of Molecular Biology, Cambridge, UK
Could you please tell us about your research field and your experience in microscopy?
My lab works on asymmetric cell division, which is the process by which one cell divides into two daughter cells with different fates. Asymmetric cell division is a fundamental process of life, in a lot of species, life starts with asymmetric cell division, and then it is responsible for the great diversity of cell types found in our bodies as it is what stem cells do. More specifically, we are interested in understanding at the molecular level how the cytoskeleton orchestrates the asymmetric segregation of the cell fate determinants in only one of the two daughter cells during asymmetric division.
We perform our studies at multiple scales (tissue, cells in culture, in vitro reconstitution with pure proteins) and thus we have experience with many different microscopy modalities (different spinning disk systems, azimuthal TIRFM, light sheet). We also actively develop microscopy techniques to manipulate biological systems (micropatterning) or to enable fast imaging of multiple channels simultaneously (high-speed multicolor imaging with X-Light V3).
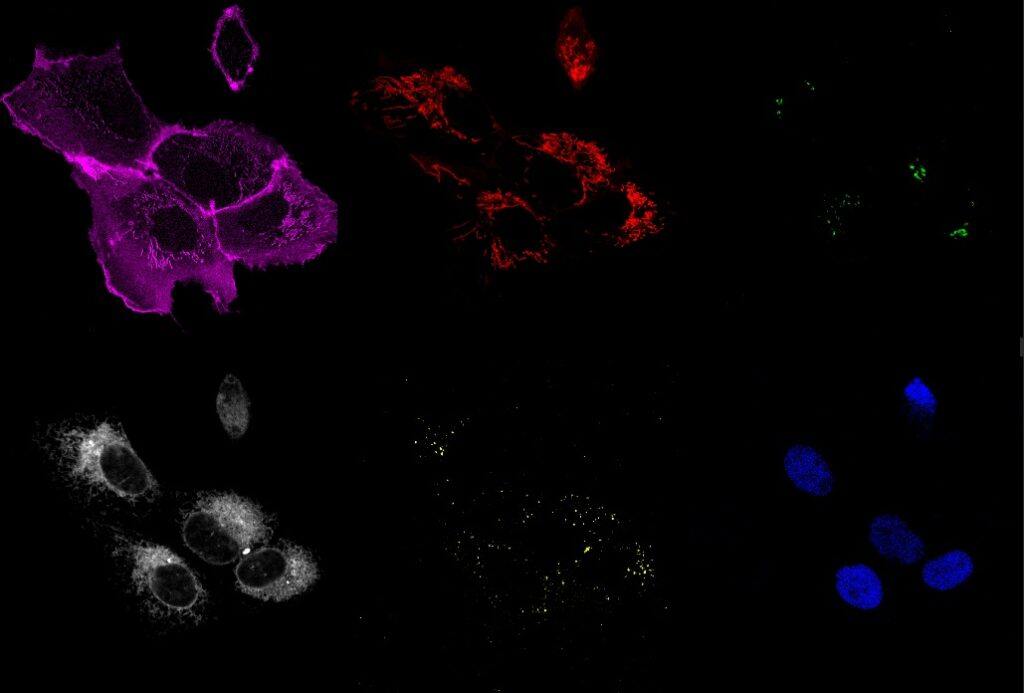
Multicolor Imaging with X-Light V3 spinning disk. A) 6-channel multicolor time-lapse acquisition U2OS cells coexpressing fluorescent proteins targetted to different organelles: Nucleus (blue; labeled with Tag BFP), Endoplasmic Reticulum (white; mCherry), Mitochondria (red; AzamiGreen); plasma membrane (Magenta; Cerulean); Golgi apparatus (green; Citrine) and Peroxisomes (yellow, labeled with iRFP670). 63X objective, time frame every 10 sec for 6 min in total. B) First time-frame showing the individual 6 channels acquired. Credits: Akaash Kumar, James Manton, and Emmanuel Derivery.
“ We perform our studies at multiple scales and thus we have experience with many different microscopy modalities”
What are the spinning disk features needed for your research?
As we transitioned from imaging single cells to imaging tissues, we sought out an instrument with a large field of view (FOV) and minimal pinhole crosstalk to allow for deep imaging without limitations. Additionally, having the capability to easily switch between widefield and confocal modes was crucial to address our biological inquiries.
“We build up a high-speed multicolor system on the X-Light V3 spinning disk with which we can perform fast live cell imaging of up to 8 channels simultaneously.”
Could you please tell us which features of the X-Light V3 spinning disk are helping your research?
We love the large field of view of the X-Light V3 coupled with its homogenous illumination. These features are relevant, for instance, for tissue imaging and spatial transcriptomics applications. Also, this spinning disk “just works”, we never had to service it, so it is perfect for a workhorse microscope everybody can use.
Moreover, the bypass mode is a very important feature for combining this head with other modalities (for instance, we are currently implementing a TIRF module on the set-up) making it a very versatile hardware for various applications and technology development.
Can you mention all the biological applications you are carrying on the CrestOptics system?
Among the applications we are interested in, I can mention: (i) imaging asymmetric cell division and cytoskeleton dynamics in fly tissues; (ii) imaging fast trafficking of receptors between different intracellular compartments monitoring multiple receptors at the same time; (iii) spatial transcriptomics in mouse tissues. For (ii) and (iii) multicolor imaging with X-Light V3 setup is required.
For multicolor imaging, we combined different fluorophore types to cover the entire spectra: we combine fluorescent proteins, organic dyes, as well as fluorescent nanobodies, along with the development of new fluorophores.
Time-lapse imaging with X-Light V3 spinning disk. Time-lapse acquisition U2OS cells expressing the actin marker LifeAct fused to the fluorescent protein mScarlet. 63X objective, time frame every 5 sec for 6 min in total. Credits: Akaash Kumar, James Manton, and Emmanuel Derivery.
How do you see the future of fluorescence microscopy to keep up with innovative biological applications and researcher requests?
I think a lot of amazing work has been done to improve spatial resolution. This opened a lot of novel avenues for biology. But comparatively, less has been done to improve temporal resolution especially when dealing with multiple fluorophores. “Life is complex”, so we need to focus on imaging the protein interactome underlying biological processes now! Similarly, very long-term imaging (days/weeks) remains a challenge, but this is the timescale of organism development, which is so important to biology but still rather understudied. For this later point, I think quantitative label-free imaging offers some amazing opportunities (for instance QPI coupled with AI).